Channels
Special Offers & Promotions
Scientists uncover a molecule that can help coronavirus escape antibodies
Researchers have found that a natural molecule can effectively block the binding of a subset of human antibodies to SARS-CoV-2. The discovery may help explain why some COVID-19 patients can become severely ill despite having high levels of antibodies against the virus.
In their research, published in Science Advances, teams from the Francis Crick Institute, in collaboration with researchers at Imperial College London, Kings College London and UCL (University College London), found that biliverdin and bilirubin, natural molecules present in the body, can suppress the binding of antibodies to the coronavirus spike.
As vaccines are rolled out globally, understanding immunity to SARS-CoV-2 and also how the virus evades antibodies is critically important. However, there are still many unknowns. The ability of the immune system to control the infection and the quality of the antibody response are highly variable, and not well correlated, between individuals.
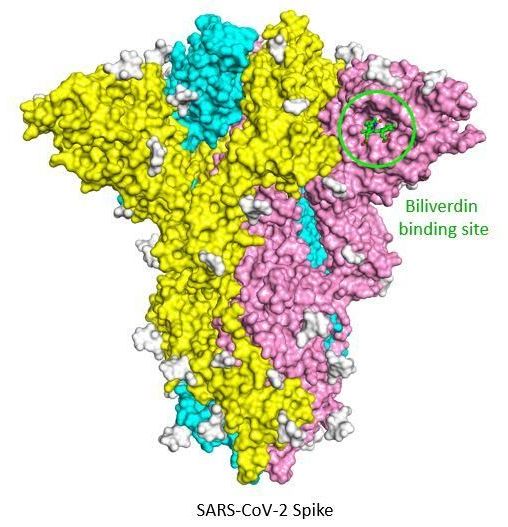
The Crick researchers were involved in the development of tests that see if a person has been exposed to the virus. The scientists discovered that the SARS-CoV-2 spike protein strongly binds to biliverdin, a molecule which was giving these proteins an unusual green colouration.
Working with teams at Imperial College London, UCL and Kings College London, they found that this natural molecule reduced antibody binding to the spike. They used blood sera and antibodies from people who were previously infected with SARS-CoV-2 and found that biliverdin could suppress the binding of human antibodies to the spike by as much as 30-50%, with some antibodies becoming ineffective at neutralising the virus.
Such a significant impact was completely unexpected, as biliverdin only binds to a very small patch on the virus' surface. To find out the mechanism at work, the team at the Crick used cryo-electron microscopy and X-ray crystallography to look in detail at the interactions between the spike, the antibodies and biliverdin. They found that biliverdin attaches to the spike N-terminal domain and stabilises it so that the spike is not able to open up and expose parts of its structure. This means that some antibodies are not able to access their target sites and so cannot bind to and neutralise the virus.
Annachiara Rosa, first author and postdoctoral training fellow in the Chromatin structure and mobile DNA Laboratory at the Crick, says: “When SARS-CoV-2 infects a patient’s lungs it damages blood vessels and causes a rise in the number immune cells. Both of these effects may contribute to increasing the levels of biliverdin and bilirubin in the surrounding tissues. And with more of these molecules available, the virus has more opportunity to hide from certain antibodies. This is a really striking process, as the virus may be benefiting from a side-effect of the damage it has already caused.”
Peter Cherepanov, author and a group leader of the Chromatin structure and mobile DNA Laboratory at the Crick, says: “In the first months of the pandemic, we were extremely busy churning out viral antigens for SARS-CoV-2 tests. It was a race, as these tests were urgently needed. When we finally found the time to study our green proteins, we expected a mundane answer. Instead, we were astonished to discover a new trick the virus uses to avoid antibody recognition. This is a result of a collaborative effort of several amazing teams working at the Crick and three partner universities, led purely by scientific curiosity.”
The researchers will continue this work from various angles, including measuring the levels of biliverdin and other haem metabolites in patients with COVID-19 and also exploring if it is possible to hijack the binding site used by biliverdin to potentially find new ways to target the virus.
Media Partners