Members Login

Channels
Special Offers & Promotions
ChIP-seq Assay Kit for Histone Methylation
Chromatrap® reports on the advantages of using its Chromatin Immunoprecipitation Sequencing (ChIP-seq) assay kits for histone methylation applications.
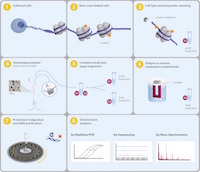 Chromatrap’s® ChIP-seq kit uses solid state technology in parallel with high throughput sequencing to deliver a streamlined ChIP-seq protocol from small cell numbers and low chromatin concentrations to investigate the epigenetic landscape. Specifically adapted for broader chromatin concentrations, Chromatrap® ChIP-seq combines the dynamic range of Chromatrap® with the downstream analysis power of deep sequencing. This allows faster, more reproducible genome wide identification of histone methylation sites. With no limitation in scale or resolution, Next Generation Sequencing can elucidate the role of epigenetic marks on gene transcription and epigenetic chromatin status.
Chromatrap’s® ChIP-seq kit uses solid state technology in parallel with high throughput sequencing to deliver a streamlined ChIP-seq protocol from small cell numbers and low chromatin concentrations to investigate the epigenetic landscape. Specifically adapted for broader chromatin concentrations, Chromatrap® ChIP-seq combines the dynamic range of Chromatrap® with the downstream analysis power of deep sequencing. This allows faster, more reproducible genome wide identification of histone methylation sites. With no limitation in scale or resolution, Next Generation Sequencing can elucidate the role of epigenetic marks on gene transcription and epigenetic chromatin status.
The Chromatrap® ChIP-seq kit allows the user to perform up to 24 ChIP assays from cell collection through to immunoprecipitation, including up to 10 chromatin sample preparations. The kit provides all of the major components required for performing ChIP assays to obtain high quality DNA for Next Generation Sequencing library preparation. With the Chromatrap® ChIP-seq kit you can sequence from as little as 1-50 µg of chromatin and perform up to 10 library preparations form a single IP. A complete ChIP-seq assay can be completed in just two days.
Launched worldwide in 2012 - Chromatrap® solid-state ChIP technology has been shown by a growing number of research groups worldwide to be more efficient than conventional bead-based methods. This is because the solid phase porous polymer, functionalized with either protein A or G, provides a greater surface area for chromatin antibody binding with very low non-specific binding. In addition, it uses a spin column approach, offering significant speed, process and carry-over advantages over sepharose or magnetic beads. DNA pull down with Chromatrap® is up to 25 times more than conventional methods, whilst the signal to noise ratio for DNA enrichment is 2 to 3 times better, even with low chromatin samples between 50ng to 3000ng per immunoprecipitation.
Media Partners