Channels
Special Offers & Promotions
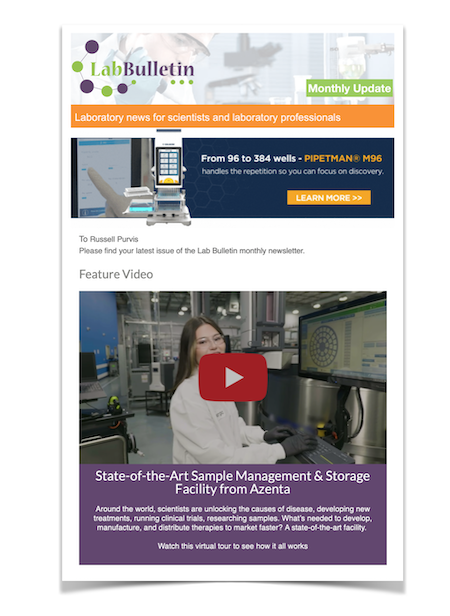
High throughput clinical and environmental genome sequencing for COVID-19
Dr Christopher Mason, Professor of Genomics, Physiology and Biophysics at Weill Cornell Medicine.
At the start of the COVID-19 pandemic in March 2020, Dr Mason and his team set about turning their knowledge of functional genomics to sequencing and understanding the clinical impact of the SARS-CoV-2 virus. The lab developed multiple methods to detect infections, track strain evolution, and identify biomarkers of disease course to assist with clinical identification and treatment. They also applied their experience in environmental genome monitoring to track the presence of the virus.
Identification and sequencing of SARS-CoV-2
With the need to quickly gain a better understanding of the spread of the SARS-CoV-2 virus – and its impact on patients – the Mason group carried out high throughput RNA and DNA extraction from naso-oropharyngeal swabs collected from the first 735 potential COVID-19 patients at the New York-Presbyterian Hospital-Weill Cornell Medicine. They then performed qRT-PCR and LAMP analysis for SARS-CoV-2 infection, identifying 216 positive patients. The team also carried out large-scale shotgun metatranscriptomics (total-RNA-seq) for host, viral and microbial profiling, using the data to develop a broad molecular portrait of the emerging COVID-19 disease. Sequencing of the virus and comparison to the Wuhan strain found significant enrichment of a NYC-distinctive clade of the virus (20C)1.
Clinical implication of SARS-CoV-2 infection
The group next looked for correlations between COVID-19 infection and changes in the microbiome and host gene expression, noting that the microbiome of the upper respiratory tract was disrupted in patients with high SARS-CoV-2 titres. However, whether this disruption was a result of the infection, or a cause for the risk of the infection, remains to be determined with longitudinal sampling. Looking at differential gene expression of human genes identified upregulation of interferon response to infection, as expected. More surprisingly, other networks of genes were affected, including downregulation of the haemoglobin complex and olfactory receptors – which may explain the loss of smell and taste often experienced by COVID-19 patients. The data also validated the role of complement and coagulation factors in clotting disorders identified in COVID-19 patients2.
Environmental viral monitoring
The Mason lab used the same methodology to swab different surfaces in various hospitals to monitor for the presence of the virus3. The hospital was divided in to 3 zones: ‘hot zones’ – where there were patients with active infections – ‘warm zones’ and ‘cold zones’, which were geographically close to or far away from the hot zones, respectively. As expected, wall swabs in hot zones detected the presence of the virus, and levels were reduced in both warm and cold zones. Interestingly, analyses of ‘high tough’ surfaces in cold zones – such as toilet handles and workstations – still showed detectable levels of virus, suggesting that staff or patients were carrying the virus out of the hot zone. They also detected virus on the ceiling above toilets in both hot and cold zones, suggesting aerosolization of the virus from flushing toilets.
As part of the global Metagenomics & Metadesign of Subways & Urban Biomes (MetaSUB) project, the group also tracked the presence of the virus in swabs collected by coordinated sampling of subways and hospitals in 59 cities across 25 countries. SARS-CoV-2 virus could be identified on subway surfaces in many cities, and its relative abundance corresponded with the severity of the outbreak in the area at the time, suggesting that environmental viral monitoring offers an opportunity to track outbreaks.
Beyond COVID-19
The approaches developed by the Mason group and its collaborators are not limited to SARS-CoV-2, and indeed can be used to study a wide range of microorganisms. For example, nearly 50 % of the DNA from samples collected from every station on the New York subway was from unknown organisms – highlighting the vast world of microbes whose biology remains unstudied4. The MetaSUB project team chose to analyse the other RNA and DNA present in the subway swabs, and sequencing has led to an increase of more than three-fold in the size of the tree of life, including the identification of 10,928 entirely novel viruses and hundreds of new bacteria5,6. From these studies, the group have also built maps of novel peptides and novel CRISPR arrays, developing the starting block for a planet-wide genetic map, which also has implications for work on Planetary Protection7 and also forensic applications8.
More information on the groups exploration of the global genome can be found in Christopher’s talk at the recent Tecan Genomics Symposium - here.
Conclusions
The COVID-19 pandemic has demonstrated that combination of high throughput RNA/DNA extraction and shotgun whole genome sequencing has clear clinical applications to monitor viruses, and led to multiple emergency use authorizations by the FDA. These tools allow monitoring of mutations and provides insights into the biology of the virus and its impact on its host. Beyond clinical applications, environmental genome sequencing provides the opportunity to track the presence of pathogens around the globe, and demonstrates the vast world of microbes that remain understudied.
References
1 - Butler, D., Mozsary, C., Meydan, C. et al. Shotgun transcriptome, spatial omics, and isothermal profiling of SARS-CoV-2 infection reveals unique host responses, viral diversification, and drug interactions. Nat Commun 12, 1660 (2021). https://doi.org/10.1038/s41467-021-21361-7
2 - Ramlall, V., Thangaraj, P.M., Meydan, C. et al. Immune complement and coagulation dysfunction in adverse outcomes of SARS-CoV-2 infection. Nat Med 26, 1609–1615 (2020). https://doi.org/10.1038/s41591-020-1021-2
3 - Brune Z, Kuschner CE, Mootz J, et al. Effectiveness of SARS-CoV-2 Decontamination and Containment in a COVID-19 ICU. Int. J. Environ. Res. Public Health, 18(5), 2479 (2021). https://doi.org/10.3390/ijerph18052479
4 - Afshinnekoo, E., Meydan, C., Chowdhury S., et al. Geospatial Resolution of Human and Bacterial Diversity with City-Scale Metagenomics. Cell Systems, Volume 1, Issue 1, 72-87 (2015). https://doi.org/10.1016/j.cels.2015.01.001
5 - Parks, D.H., Rinke, C., Chuvochina, M. et al. Recovery of nearly 8,000 metagenome-assembled genomes substantially expands the tree of life. Nat Microbiol 2, 1533–1542 (2017). https://doi.org/10.1038/s41564-017-0012-7
6 - Danko D.C., Bezdan D., Afshinnekoo E., et al. A global metagenomic map of urban microbiomes and antimicrobial resistance Cell (2021). https://doi.org/10.1016/j.cell.2021.05.002
7 – Danko D.C., Sierra M.A., Benardini J.N., et al. A comprehensive metagenomics framework to characterize organisms relevant for planetary protection. Microbiome 9, 82 (2021). https://doi.org/10.1186/s40168-021-01020-1
8 - Elhaik E, Ahsanuddin S, et al. The impact of cross-kingdom molecular forensics on genetic privacy. Microbiome 9, 114 (2021). https://doi.org/10.1186/s40168-021-01076-z
Media Partners